Regime changes 3.5 - malignantventricular with PR
Francisco Bischoff
on August 13, 2023
Last updated: 2023-08-13
Checks: 7 0
Knit directory:
false.alarm/docs/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20201020) was run prior to running the code in the R Markdown file.
Setting a seed ensures that any results that rely on randomness, e.g.
subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 4437e7a. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the
analysis have been committed to Git prior to generating the results (you can
use wflow_publish or wflow_git_commit). workflowr only
checks the R Markdown file, but you know if there are other scripts or data
files that it depends on. Below is the status of the Git repository when the
results were generated:
Ignored files:
Ignored: .Renviron
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: .devcontainer/exts/
Ignored: .docker/
Ignored: .github/ISSUE_TEMPLATE/
Ignored: .httr-oauth
Ignored: R/RcppExports.R
Ignored: _classifier/meta/process
Ignored: _classifier/meta/progress
Ignored: _classifier/objects/
Ignored: _classifier/user/
Ignored: _contrast_profile/meta/process
Ignored: _contrast_profile/meta/progress
Ignored: _contrast_profile/objects/
Ignored: _contrast_profile/user/
Ignored: _contrast_profile_ex/meta/process
Ignored: _contrast_profile_ex/meta/progress
Ignored: _contrast_profile_ex/objects/
Ignored: _contrast_profile_ex/user/
Ignored: _contrast_profile_ex/workspaces/
Ignored: _regime_change/meta/process
Ignored: _regime_change/meta/progress
Ignored: _regime_change/objects/
Ignored: _regime_change/user/
Ignored: _regime_change2/meta/process
Ignored: _regime_change2/meta/progress
Ignored: _regime_change2/objects/
Ignored: _regime_change2/user/
Ignored: _regime_change3/meta/process
Ignored: _regime_change3/meta/progress
Ignored: _regime_change3/objects/
Ignored: _regime_change3/user/
Ignored: _regime_change_2/meta/process
Ignored: _regime_change_2/meta/progress
Ignored: _regime_change_2/objects/
Ignored: _regime_change_2/user/
Ignored: _regime_optimize/meta/meta2
Ignored: _regime_optimize/meta/process
Ignored: _regime_optimize/meta/progress
Ignored: _regime_optimize/objects/
Ignored: _regime_optimize/user/
Ignored: _targets/meta/process
Ignored: _targets/meta/progress
Ignored: _targets/objects/
Ignored: _targets/user/
Ignored: analysis/report_cache/
Ignored: analysis/shiny/rsconnect/
Ignored: analysis/shiny_land/rsconnect/
Ignored: analysis/shiny_ventricular/rsconnect/
Ignored: analysis/shiny_vtachy/rsconnect/
Ignored: dev/
Ignored: inst/extdata/
Ignored: output/importancess.rds
Ignored: papers/aime2021/aime2021.md
Ignored: papers/epia2022/epia2022.md
Ignored: presentations/MEDCIDS21/MEDCIDS21-10min_files/
Ignored: presentations/MEDCIDS21/MEDCIDS21_files/
Ignored: presentations/Report/Midterm-Report_cache/
Ignored: presentations/Report/Midterm-Report_files/
Ignored: protocol/SecondReport_cache/
Ignored: protocol/SecondReport_files/
Ignored: protocol/ThirdReport.tex
Ignored: protocol/ThirdReport_cache/
Ignored: protocol/ThirdReport_files/
Ignored: protocol/_extensions/
Ignored: protocol/_files/
Ignored: protocol/figure/
Ignored: renv/staging/
Ignored: src/RcppExports.cpp
Ignored: src/RcppExports.o
Ignored: src/contrast.o
Ignored: src/false.alarm.so
Ignored: src/fft.o
Ignored: src/mass.o
Ignored: src/math.o
Ignored: src/mpx.o
Ignored: src/scrimp.o
Ignored: src/stamp.o
Ignored: src/stomp.o
Ignored: src/windowfunc.o
Ignored: thesis/Rplots.pdf
Ignored: thesis/_bookdown_files/
Ignored: tmp/
Unstaged changes:
Modified: analysis/regime_optimize.Rmd
Modified: analysis/report.Rmd
Modified: output/importances.rds
Modified: output/importances2.rds
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made
to the R Markdown (analysis/regime_optimize_3_5.Rmd) and HTML (docs/regime_optimize_3_5.html)
files. If you’ve configured a remote Git repository (see
?wflow_git_remote), click on the hyperlinks in the table below to
view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d06c64f | Francisco Bischoff | 2022-10-09 | Squashed commit of the following: |
| html | d06c64f | Francisco Bischoff | 2022-10-09 | Squashed commit of the following: |
| html | f9f551d | Francisco Bischoff | 2022-10-06 | Build site. |
| Rmd | c052902 | Francisco Bischoff | 2022-10-06 | another score |
1 Regime changes optimization (continuation)
This article uses the same principles as the previous article but here we will evaluate the models in another dataset, the “CU Ventricular Tachyarrhythmia Database” which contains 35 eight-minute ECG recordings of human subjects who experienced episodes of sustained ventricular tachycardia, ventricular flutter, and ventricular fibrillation.
This version of the analysis uses another scoring function, which is based on the F-score, and uses a simple approach to calculate the Precision and Recall. The Recall reflects the hability of the model to detect the labeled regime changes; the algorithm accounts as a “detection” if the prediction is within a window of 5 seconds (plus or minus 2.5s). The Precision reflects the hability of the model to avoid false positives; the algorithm accounts as a “false positive” when the prediction is outside a window of 10 seconds (plus or minus 5s) from the labeled regime change.
The F-score will be calculated giving more weight to the Recall (beta = 3). Except for the Fig. 1.9, all other figures will represent the F-score as \(Score = 1 - F_{score}\), to make it easier to compare with the previous score function.
1.1 Current pipeline
Figure 1.1: FLOSS pipeline.
1.2 Tuning process
This time, as we have already seen the results of the previous optimization, we will only tune the parameters that we have concluded that they are meaningful.
The variable for building the MP:
window_size: the default parameter always used to build an MP.
The variables used on the FLOSS algorithm:
regime_threshold: the threshold below which a regime change is considered.regime_landmark: the point in time where the regime threshold is applied.
Using the tidymodels framework, we performed a basic grid search on all these parameters as follows:
- The MP parameters were explored using the following values:
window_size: 25 to 200, by 25 steps;
- The FLOSS parameters were explored using the following values:
regime_threshold: 0.05 to 0.90, by 0.05 steps;regime_landmark: 2 to 9.5, by 0.5 steps.
1.3 Parameters analysis
As before, we started by computing the importance of each parameter1. We used the same approach using the Bayesian Additive Regression Trees (BART) model to fit the tuning parameters as predictors of the FLOSS score.
1.3.1 Interactions
Before starting the parameter importance analysis, we need to consider the parameter interactions since this is usually the weak spot of the analysis techniques.
The BART model was fitted using the following parameters:
\[\begin{equation} \begin{aligned} E( score ) &= \alpha + window\_size\\ &\quad + regime\_threshold + regime\_landmark \end{aligned} \tag{1.1} \end{equation}\]
Fig. 1.2 shows the variable interaction strength between pairs of variables. That allows us to verify if there are any significant interactions between the variables. Here we see that the interaction between variables are minimal.
Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.
This warning is displayed once every 8 hours.
Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
generated.Figure 1.2: Variable interactions strength using feature importance ranking measure (FIRM) approach2. Shows almost no interaction between the variables.
1.3.2 Importance
After evaluating the interactions, we can then perform the analysis of the variable importance. The goal is to understand how the FLOSS score behaves when we change the parameters.
The techniques for evaluating the variable importances were described in the previous article.
1.3.3 Importance analysis
Using the three techniques simultaneously allows a broad comparison of the model behavior3. All three methods are model-agnostic (separates interpretation from the model), but as we have seen, each method has its advantages and disadvantages4.
Fig. 1.3 then shows the variable importance using three methods: Feature Importance Ranking Measure (FIRM) using Individual Conditional Expectation (ICE), Permutation-based, and Shapley Additive explanations (SHAP). Here we see that all three methods agree on the importances.
Figure 1.3: Variables importances using three different methods. A) Feature Importance Ranking Measure using ICE curves. B) Permutation method. C) SHAP (400 iterations).
Fig. 1.4 shows the effect of each feature on the FLOSS score.
Based on Figures 1.3 and 1.4 we can infer that:
regime_threshold: is the most important feature, as in the previous dataset, and have a converging value.regime_landmark: seems to have lost its importance using this metric.window_size: has some importance, and as before, value below 75 starts to degrade the FLOSS score.
Figure 1.4: This shows the effect each variable has on the FLOSS score. This plot doesn’t take into account the variable interactions.
1.4 Visualizing the predictions
1.4.1 By recording
First, we will visualize how the models (in general) performed throughout the individual recordings.
Fig. 1.5 shows a violin plot of equal areas clipped to the minimum value. The blue color indicates the recordings with a small IQR (interquartile range) of model scores. We see on the left half 50% of the recordings with the worst minimum score, and on the right half, 50% of the recordings with the best minimum score.
Next, we will visualize some of these predictions to understand why some recordings were difficult to segment.
Warning: Returning more (or less) than 1 row per `summarise()` group was deprecated in
dplyr 1.1.0.
ℹ Please use `reframe()` instead.
ℹ When switching from `summarise()` to `reframe()`, remember that `reframe()`
always returns an ungrouped data frame and adjust accordingly.
Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
generated.Figure 1.5: Violin plot showing the distribution of the FLOSS score achieved by all tested models by recording. The left half shows the recordings that were difficult to predict (50% overall), whereas the right half shows the recordings that at least one model could achieve a good prediction (50% overall). The recordings are sorted (left-right) by the minimum (best) score achieved in descending order, and ties are sorted by the median of all recording scores. The blue color highlights recordings where models had an IQR variability of less than 0.5.
Fig. 1.6 shows the best effort in predicting the most complex recordings. One information not declared before is that if the model does not predict any change, it will put a mark on the zero position. On the other side, the truth markers positioned at the beginning and the end of the recording were removed, as these locations lack information and do not represent a streaming setting.
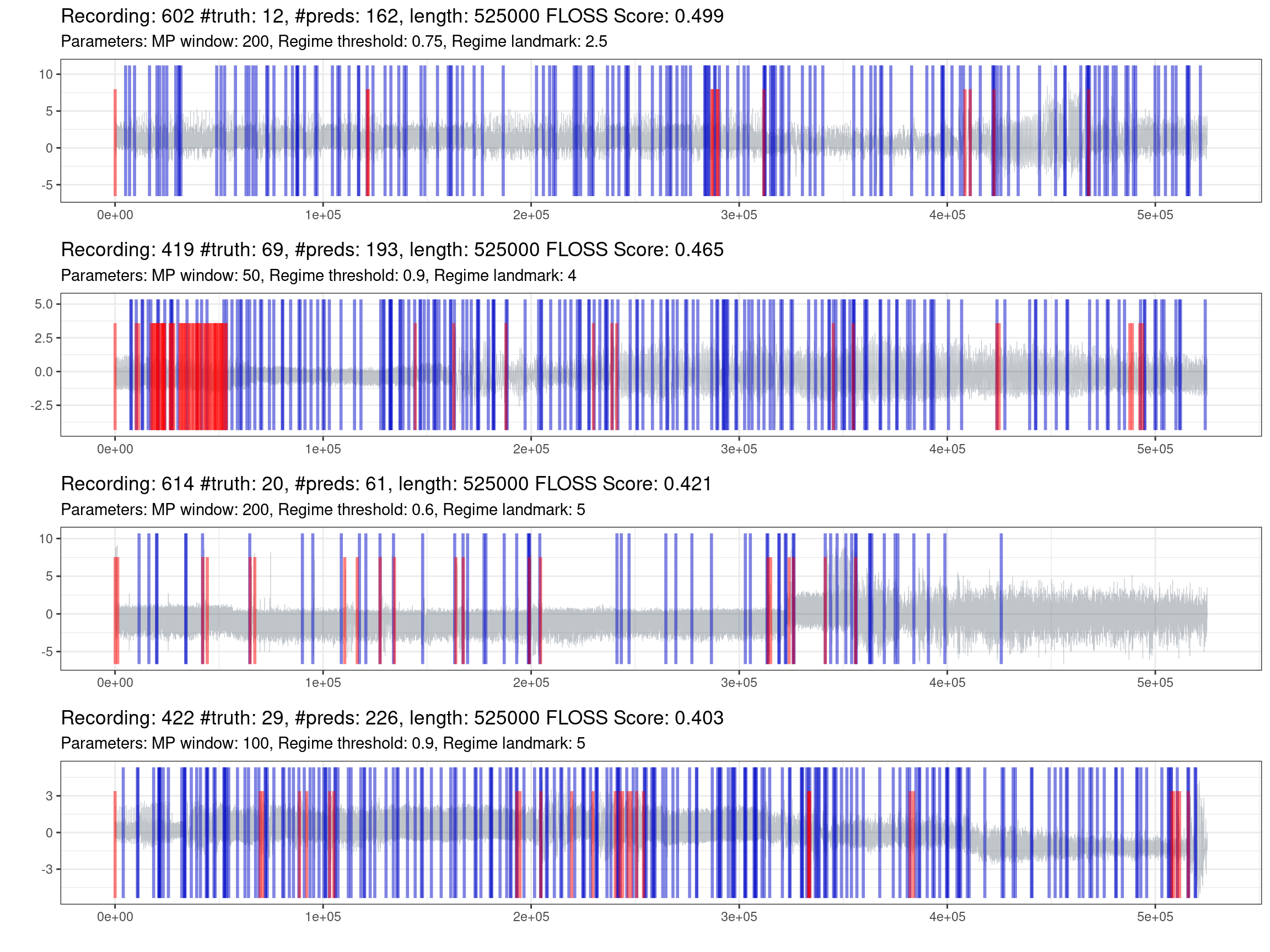
Figure 1.6: Prediction of the worst recordings (red is the truth, blue are the predictions).
Fig. 1.7 shows the best performances of the best recordings.
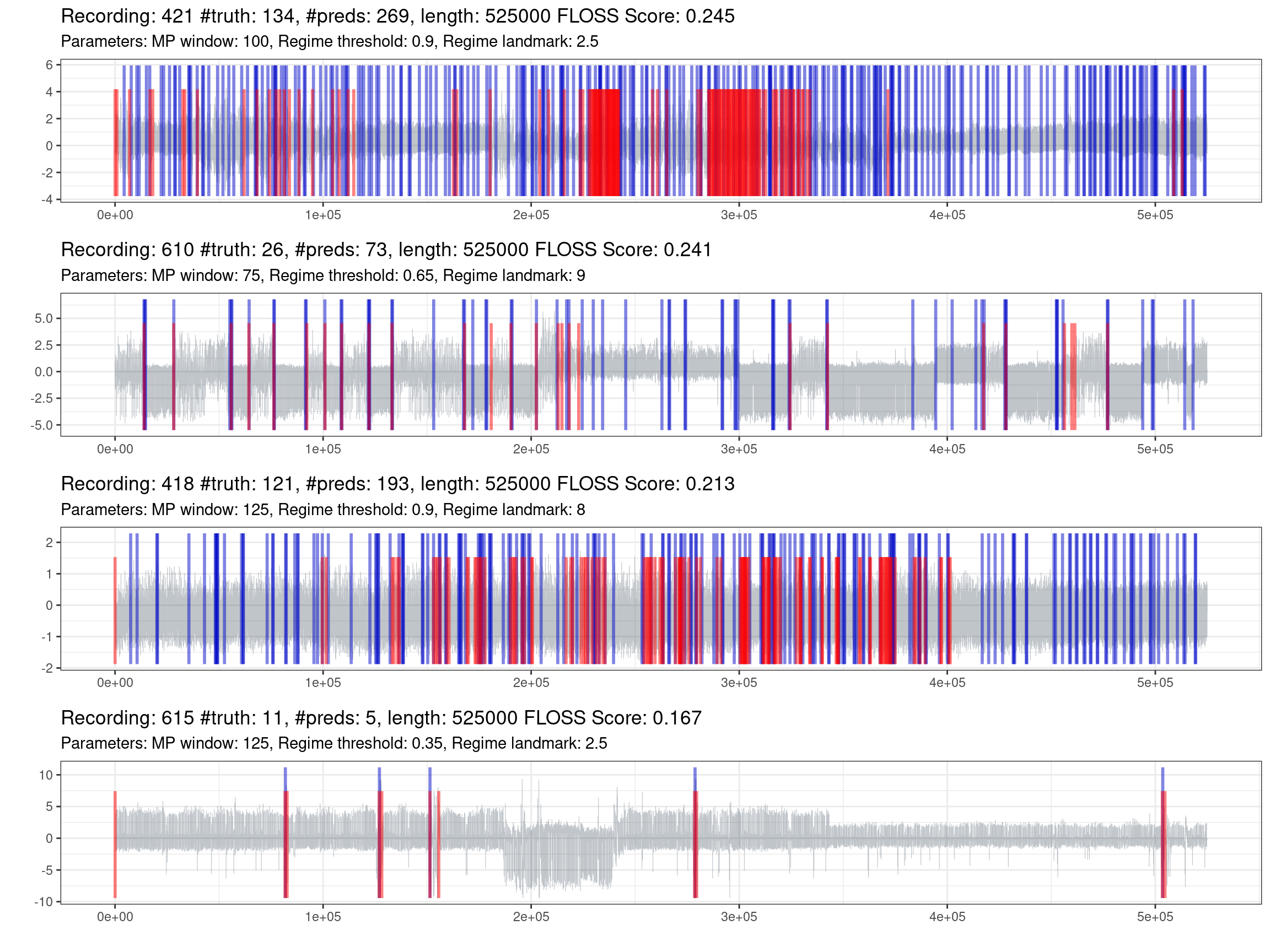
Figure 1.7: Prediction of the best recordings (red is the truth, blue are the predictions).
An online interactive version of all the datasets and predictions can be accessed at Shiny app (default Score).
1.4.2 By model
Fig. 1.8 shows the distribution of the FLOSS score of the 25% worst (left side) and 25% best models across the recordings (right side). The bluish color highlights the models with SD below 0.5 and IQR below 0.5.
Warning: Returning more (or less) than 1 row per `summarise()` group was deprecated in
dplyr 1.1.0.
ℹ Please use `reframe()` instead.
ℹ When switching from `summarise()` to `reframe()`, remember that `reframe()`
always returns an ungrouped data frame and adjust accordingly.
Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
generated.Figure 1.8: Violin plot showing the distribution of the FLOSS score achieved by all tested models during the inner ressample. The left half shows the models with the worst performances (top 25), whereas the right half shows the models with the best performances (top 25). The models are sorted (left-right) by the mean score (top) and by the median (below). Ties are sorted by the SD and IQR, respectively. The bluish colors highlights models with an SD below 0.5 and IQR below 0.5.
Fig. 1.9 the performance of the six best models. They are ordered from left to right, from the best record to the worst record. The top model is the one with the highest mean across the scores. The blue line indicates the precision of the model, and the green line the recall. The black line with dots is the f-score. The red continuous and dashed lines are the median and the mean f-score respectively. The gray line limits the zero-score region.
Figure 1.9: Performances of the best 6 models across all inner resample of recordings. The recordings are ordered by score, from the best to the worst. Each plot shows one model, starting from the best one. The blue line indicates the precision of the model. The green line the recall. The black line with dots is the f-score. The red continuous and dashed lines indicate respectively the median and mean f-score of the model. The gray line limits the zero-score region.
References
─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.1 (2023-06-16)
os Ubuntu 22.04.2 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz Europe/Lisbon
date 2023-08-13
pandoc 2.17.0.1 @ /usr/bin/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
askpass 1.1 2019-01-13 [1] CRAN (R 4.3.0)
backports 1.4.1 2021-12-13 [1] CRAN (R 4.3.1)
base64url 1.4 2018-05-14 [1] CRAN (R 4.3.0)
bit 4.0.5 2022-11-15 [1] CRAN (R 4.3.0)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.3.0)
bookdown 0.35.1 2023-08-13 [1] Github (rstudio/bookdown@661567e)
bslib 0.5.1 2023-08-11 [1] CRAN (R 4.3.1)
cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.0)
callr 3.7.3 2022-11-02 [1] CRAN (R 4.3.1)
checkmate 2.2.0 2023-04-27 [1] CRAN (R 4.3.0)
class 7.3-22 2023-05-03 [2] CRAN (R 4.3.1)
cli 3.6.1 2023-03-23 [1] CRAN (R 4.3.1)
codetools 0.2-19 2023-02-01 [2] CRAN (R 4.3.0)
colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
crayon 1.5.2 2022-09-29 [1] CRAN (R 4.3.1)
credentials 1.3.2 2021-11-29 [1] CRAN (R 4.3.0)
data.table 1.14.8 2023-02-17 [1] CRAN (R 4.3.0)
dbarts 0.9-23 2023-01-23 [1] CRAN (R 4.3.0)
debugme 1.1.0 2017-10-22 [1] CRAN (R 4.3.0)
devtools 2.4.5 2022-10-11 [1] CRAN (R 4.3.0)
dials 1.2.0 2023-04-03 [1] CRAN (R 4.3.0)
DiceDesign 1.9 2021-02-13 [1] CRAN (R 4.3.0)
digest 0.6.33 2023-07-07 [1] CRAN (R 4.3.1)
dplyr 1.1.2 2023-04-20 [1] CRAN (R 4.3.0)
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.3.0)
evaluate 0.21 2023-05-05 [1] CRAN (R 4.3.0)
fansi 1.0.4 2023-01-22 [1] CRAN (R 4.3.0)
farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
fastshap 0.0.7 2021-12-06 [1] CRAN (R 4.3.0)
forcats 1.0.0 2023-01-29 [1] CRAN (R 4.3.0)
foreach 1.5.2 2022-02-02 [1] CRAN (R 4.3.0)
fs 1.6.3 2023-07-20 [1] CRAN (R 4.3.1)
furrr 0.3.1 2022-08-15 [1] CRAN (R 4.3.0)
future 1.33.0 2023-07-01 [1] CRAN (R 4.3.1)
future.apply 1.11.0 2023-05-21 [1] CRAN (R 4.3.1)
future.callr 0.8.2 2023-08-09 [1] CRAN (R 4.3.1)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
gert 1.9.3 2023-08-07 [1] CRAN (R 4.3.1)
getPass 0.2-2 2017-07-21 [1] CRAN (R 4.3.0)
ggplot2 * 3.4.2 2023-04-03 [1] CRAN (R 4.3.0)
git2r 0.32.0.9000 2023-06-30 [1] Github (ropensci/git2r@9c42d41)
gittargets * 0.0.6.9000 2023-05-05 [1] Github (wlandau/gittargets@2d448ff)
globals 0.16.2 2022-11-21 [1] CRAN (R 4.3.0)
glue * 1.6.2 2022-02-24 [1] CRAN (R 4.3.1)
gower 1.0.1 2022-12-22 [1] CRAN (R 4.3.0)
GPfit 1.0-8 2019-02-08 [1] CRAN (R 4.3.0)
gridExtra 2.3 2017-09-09 [1] CRAN (R 4.3.0)
gtable 0.3.3 2023-03-21 [1] CRAN (R 4.3.0)
hardhat 1.3.0 2023-03-30 [1] CRAN (R 4.3.0)
here * 1.0.1 2020-12-13 [1] CRAN (R 4.3.0)
highr 0.10 2022-12-22 [1] CRAN (R 4.3.1)
hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.0)
htmltools 0.5.6 2023-08-10 [1] CRAN (R 4.3.1)
htmlwidgets 1.6.2 2023-03-17 [1] CRAN (R 4.3.0)
httpuv 1.6.11 2023-05-11 [1] CRAN (R 4.3.1)
httr 1.4.6 2023-05-08 [1] CRAN (R 4.3.1)
igraph 1.5.1 2023-08-10 [1] CRAN (R 4.3.1)
ipred 0.9-14 2023-03-09 [1] CRAN (R 4.3.0)
iterators 1.0.14 2022-02-05 [1] CRAN (R 4.3.0)
jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.3.0)
jsonlite 1.8.7 2023-06-29 [1] CRAN (R 4.3.0)
kableExtra * 1.3.4 2021-02-20 [1] CRAN (R 4.3.0)
knitr 1.43 2023-05-25 [1] CRAN (R 4.3.0)
labeling 0.4.2 2020-10-20 [1] CRAN (R 4.3.0)
later 1.3.1 2023-05-02 [1] CRAN (R 4.3.1)
lattice 0.21-8 2023-04-05 [2] CRAN (R 4.3.0)
lava 1.7.2.1 2023-02-27 [1] CRAN (R 4.3.0)
lhs 1.1.6 2022-12-17 [1] CRAN (R 4.3.0)
lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.3.1)
listenv 0.9.0 2022-12-16 [1] CRAN (R 4.3.0)
lubridate 1.9.2 2023-02-10 [1] CRAN (R 4.3.0)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.1)
MASS 7.3-60 2023-05-04 [2] CRAN (R 4.3.1)
Matrix 1.6-0 2023-07-08 [1] CRAN (R 4.3.1)
memoise 2.0.1 2021-11-26 [1] CRAN (R 4.3.0)
mgcv 1.9-0 2023-07-11 [1] CRAN (R 4.3.1)
mime 0.12 2021-09-28 [1] CRAN (R 4.3.0)
miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 4.3.0)
modelenv 0.1.1 2023-03-08 [1] CRAN (R 4.3.0)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.0)
nlme 3.1-163 2023-08-09 [1] CRAN (R 4.3.1)
nnet 7.3-19 2023-05-03 [2] CRAN (R 4.3.1)
openssl 2.1.0 2023-07-15 [1] CRAN (R 4.3.1)
parallelly 1.36.0 2023-05-26 [1] CRAN (R 4.3.1)
parsnip 1.1.0 2023-04-12 [1] CRAN (R 4.3.0)
patchwork * 1.1.2 2022-08-19 [1] CRAN (R 4.3.0)
pdp 0.8.1 2023-06-22 [1] Github (bgreenwell/pdp@4f22141)
pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
pkgbuild 1.4.2 2023-06-26 [1] CRAN (R 4.3.1)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
pkgload 1.3.2.1 2023-07-08 [1] CRAN (R 4.3.1)
prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.3.0)
processx 3.8.2 2023-06-30 [1] CRAN (R 4.3.1)
prodlim 2023.03.31 2023-04-02 [1] CRAN (R 4.3.0)
profvis 0.3.8 2023-05-02 [1] CRAN (R 4.3.1)
promises 1.2.1 2023-08-10 [1] CRAN (R 4.3.1)
ps 1.7.5 2023-04-18 [1] CRAN (R 4.3.1)
purrr 1.0.2 2023-08-10 [1] CRAN (R 4.3.1)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.1)
Rcpp 1.0.11 2023-07-06 [1] CRAN (R 4.3.1)
readr 2.1.4 2023-02-10 [1] CRAN (R 4.3.0)
recipes 1.0.7 2023-08-10 [1] CRAN (R 4.3.1)
remotes 2.4.2.1 2023-07-18 [1] CRAN (R 4.3.1)
renv 0.17.3 2023-04-06 [1] CRAN (R 4.3.1)
rlang 1.1.1 2023-04-28 [1] CRAN (R 4.3.0)
rmarkdown 2.23.4 2023-08-13 [1] Github (rstudio/rmarkdown@054d735)
rpart 4.1.19 2022-10-21 [2] CRAN (R 4.3.0)
rprojroot 2.0.3 2022-04-02 [1] CRAN (R 4.3.1)
rsample 1.1.1 2022-12-07 [1] CRAN (R 4.3.0)
rstudioapi 0.15.0 2023-07-07 [1] CRAN (R 4.3.1)
rvest 1.0.3 2022-08-19 [1] CRAN (R 4.3.0)
sass 0.4.7 2023-07-15 [1] CRAN (R 4.3.1)
scales 1.2.1 2022-08-20 [1] CRAN (R 4.3.0)
sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
shapviz 0.9.1 2023-07-18 [1] CRAN (R 4.3.1)
shiny 1.7.5 2023-08-12 [1] CRAN (R 4.3.1)
signal 0.7-7 2021-05-25 [1] CRAN (R 4.3.0)
stringi 1.7.12 2023-01-11 [1] CRAN (R 4.3.1)
stringr 1.5.0 2022-12-02 [1] CRAN (R 4.3.1)
survival 3.5-5 2023-03-12 [2] CRAN (R 4.3.1)
svglite 2.1.1.9000 2023-05-05 [1] Github (r-lib/svglite@6c1d359)
sys 3.4.2 2023-05-23 [1] CRAN (R 4.3.1)
systemfonts 1.0.4 2022-02-11 [1] CRAN (R 4.3.0)
tarchetypes * 0.7.7 2023-06-15 [1] CRAN (R 4.3.1)
targets * 1.2.2 2023-08-10 [1] CRAN (R 4.3.1)
tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
tidyr 1.3.0 2023-01-24 [1] CRAN (R 4.3.0)
tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.3.0)
timechange 0.2.0 2023-01-11 [1] CRAN (R 4.3.0)
timeDate 4022.108 2023-01-07 [1] CRAN (R 4.3.0)
timetk 2.8.3 2023-03-30 [1] CRAN (R 4.3.0)
tune 1.1.1 2023-04-11 [1] CRAN (R 4.3.0)
tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.1)
urlchecker 1.0.1 2021-11-30 [1] CRAN (R 4.3.0)
usethis 2.2.2.9000 2023-07-17 [1] Github (r-lib/usethis@467ff57)
utf8 1.2.3 2023-01-31 [1] CRAN (R 4.3.0)
uuid 1.1-0 2022-04-19 [1] CRAN (R 4.3.0)
vctrs 0.6.3 2023-06-14 [1] CRAN (R 4.3.1)
vip 0.4.0 2023-07-19 [1] CRAN (R 4.3.1)
viridisLite 0.4.2 2023-05-02 [1] CRAN (R 4.3.1)
visNetwork * 2.1.2 2022-09-29 [1] CRAN (R 4.3.0)
vroom 1.6.3 2023-04-28 [1] CRAN (R 4.3.1)
webshot 0.5.5 2023-06-26 [1] CRAN (R 4.3.1)
whisker 0.4.1 2022-12-05 [1] CRAN (R 4.3.0)
withr 2.5.0 2022-03-03 [1] CRAN (R 4.3.1)
workflowr * 1.7.0 2021-12-21 [1] CRAN (R 4.3.0)
workflows 1.1.3 2023-02-22 [1] CRAN (R 4.3.0)
xfun 0.40 2023-08-09 [1] CRAN (R 4.3.1)
xgboost 1.7.5.1 2023-03-30 [1] CRAN (R 4.3.0)
xml2 1.3.5 2023-07-06 [1] CRAN (R 4.3.1)
xtable 1.8-4 2019-04-21 [1] CRAN (R 4.3.0)
xts 0.13.1 2023-04-16 [1] CRAN (R 4.3.0)
yaml 2.3.7 2023-01-23 [1] CRAN (R 4.3.1)
yardstick 1.0.0.9000 2023-05-25 [1] Github (tidymodels/yardstick@90ab794)
zoo 1.8-12 2023-04-13 [1] CRAN (R 4.3.0)
[1] /workspace/.cache/R/renv/proj_libs/false.alarm-d6f1a0d1/R-4.3/x86_64-pc-linux-gnu
[2] /usr/lib/R/library
──────────────────────────────────────────────────────────────────────────────